In this post, I explore the Undiagnosed Diseases Network: Solving Medical Mysteries Through Team Science website.
Frequently Asked Questions
About the Undiagnosed Diseases Network
Description
The Undiagnosed Diseases Network (UDN) is a research study that is funded by the National Institutes of Health Common Fund. Its purpose is to bring together clinical and research experts from across the United States to solve the most challenging medical mysteries using advanced technologies.
Through this study, we hope to both help individual patients and families living with the burden of undiagnosed diseases, and contribute to the understanding of how the human body works.
Reports
UDN Quarterly Report – Summer 2022
UDN Quarterly Report – Spring 2022
UDN Quarterly Report – Winter 2022
UDN Quarterly Report – Fall 2021
UDN Quarterly Report – Summer 2021
UDN Quarterly Report – Spring 2021
UDN Quarterly Report – Winter 2021
UDN Quarterly Report – Fall 2020How to Apply
Participants
One of the greatest challenges faced by those living with an undiagnosed disease is the lack of information and communication. When a patient experiences symptoms that aren’t easily classifiable or seem uncommon, they can fall through the cracks of modern medicine—leaving them feeling isolated while the true cause of their disease goes unexamined.
Genes of Interest
Here, you can learn more about some of the genes we are studying.
Participant Pages
To help find patients with the same or similar condition, we are creating public web pages about participants in our study. Our goal is that healthcare providers, researchers, and families who know similar patients will find these pages. Connecting these patients with ours will ideally help us identify shared symptoms and diagnoses.
Undiagnosed Diseases Network Manual of Operations
January 15th, 2022Model Organisms
Overview
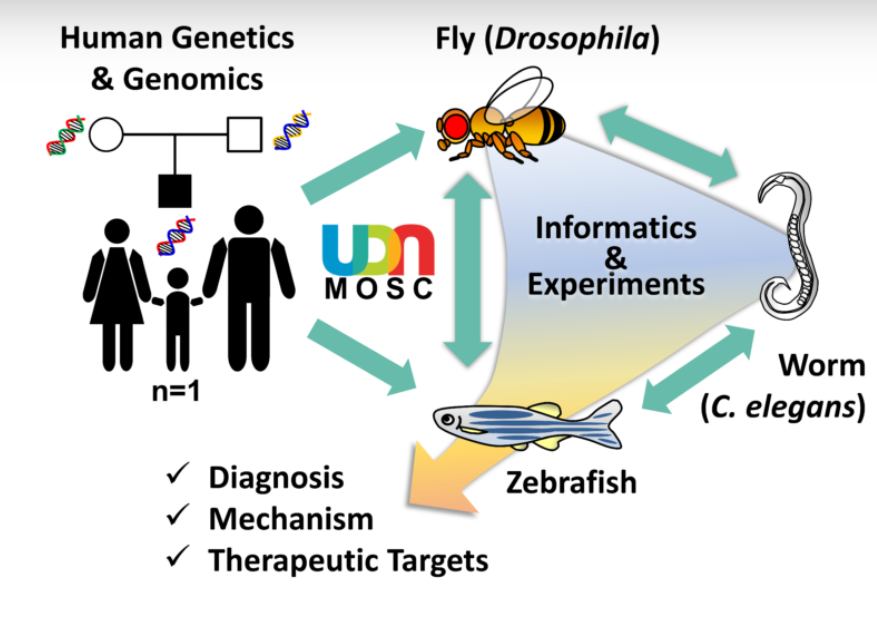
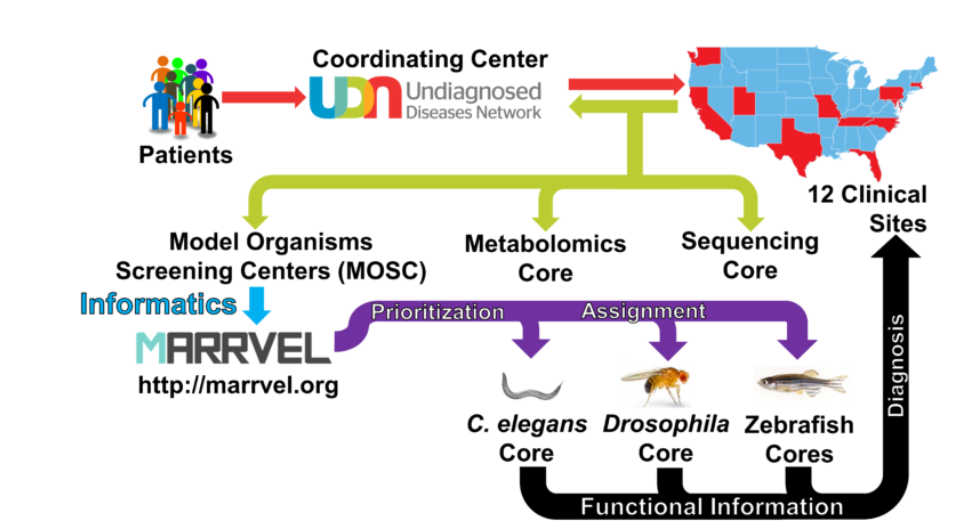
The Model Organisms Screening Center (MOSC) for the Undiagnosed Diseases Network (UDN) is composed of two Centers that use fruit fly (Drosophila melanogaster), nematode worm (Caenorhabditis elegans) and zebrafish (Danio rerio) genetics and biology to tackle rare and undiagnosed diseases. By combining state-of-the-art genetic and genomic technologies, the MOSCs investigate whether a rare variant identified in the genomes of UDN participants may contribute to disease pathogenesis. The Baylor College of Medicine (BCM)-University of Oregon (UO) MOSC is led by Drs. Hugo J. Bellen (BCM), Michael F. Wangler (BCM), Shinya Yamamoto (BCM), Monte Westerfield (UO) and John Postlethwait (UO). The Washington University in St. Louis (WUSTL) MOSC is led by Drs. Tim Schedl, Lilianna Solnica-Krezel, Dustin Baldridge, Angela Bowman and Stephen C. Pak.
Why flies, worms and zebrafish?
Over the past century, genetic model organisms have taught us so much about human biology and disease mechanisms. Although these organisms (e.g. bacteria, yeast, worm, fly, zebrafish, mouse) may look very different from us, fundamental biological mechanisms and genes are well conserved throughout evolution. To investigate the functional consequences of hundreds of rare variants found through sequencing UDN participants’ and their family members’ genomes, the MOSCs use three model organisms, fruit fly (Drosophila melanogaster), nematode worm (Caenorhabditis elegans) and zebrafish (Danio rerio). These animals are cost efficient, have short life-cycles and are amenable to sophisticated genetic manipulations to “model” a human disease condition. Drosophila, C. elegans and zebrafish are complementary to one another, providing synergistic strengths. Candidate genes and variants that are shown to have functional impacts can be further pursued in mammalian model systems, such as mouse and human pluripotent stem cells, for further translational studies.
Workflow
When a diagnosis is not reached after performing a thorough clinical, genetic and/or metabolomic workup, the UDN Clinical Sites submit candidate gene(s)/variant(s) to the MOSCs together with a brief description of the participant’s condition. The MOSCs then perform database searches using a number of bioinformatics tools, including the MARRVEL tool (marrvel.org, see below), to aggregate existing information on the human gene/variant and its model organism orthologs. The MOSCs also try to identify other individuals with similar genotype and phenotype in other cohorts, a practice known as “matchmaking”. Once a variant is considered to be a high priority candidate, experiments to assess gene and variant function are designed by MOSC investigators and pursued in the C. elegans Core, Drosophila Core or Zebrafish Cores.
MARRVEL
In collaboration with Drs. Zhandong Liu’s (BCM) and Norbert Perrimon’s (Harvard Medical School) bioinformatics team, the BCM-UO MOSC developed a powerful online tool that allows anyone to quickly gather gene and variant function information. MARRVEL (Model organism Aggregated Resources for Rare Variant ExpLoration) is a novel web-based tool that integrates human and model organism databases to facilitate molecular diagnosis. MARRVEL can also be used by model organism researchers to assess whether specific model organism genes of interest may have links to human diseases. MARRVEL is publicly available for clinicians and researchers worldwide at marrvel.org, and we will be continuously updating and upgrading this tool for the community.
Press
Mystery Cases: What Happens When Modern Medicine Lacks a Diagnosis or Cure?
June 21, 2021Discover Magazine
Nearly 100 doctors have tried to diagnose this man’s devastating illness — without success
September 21, 2019The Washington Post
They don’t know if their children will ever walk or talk. But finding other families online has given them hope.
May 6, 2019NBC News
‘Disease detectives’ crack cases of 130 patients with mysterious illnesses
October 10, 2018San Francisco Chronicle
Stanford Children’s Health helping families facing unknown diseases
October 10, 2018ABC7 News (Bay Area)
Doctors Are Becoming DNA Detectives to Diagnose Super-Rare Diseases
April 4, 2018Los Angeles Magazine
Why Fruit Flies are the New Lab Rats: These Quick-Breeding Insects Have Similar Genetic Cellular Functions as Humans
March 20, 2018Zocalo Public Square
These Houston Doctors Seek to Diagnose the Previously Un-Diagnosable
July 18, 2017Houstonia Magazine
Searching for a diagnosis: a network of doctors tries to solve medical mysteries
April 20, 2017STAT (via Kaiser Health News)
Stanford team helps patient who is “unique in the world”
December 14, 2016Scope Blog by Stanford Medicine
One of a Kind: What do you do if your child has a condition that is new to science?
July 21, 2015The New Yorker
Resources
Participant Engagement and Empowerment Resource (PEER)
The UDN Participant Engagement and Empowerment Resource (PEER) is made up of participants and family members who have participated in the UDN. UDN PEER members work with UDN researchers to improve the participant experience, connect families with each other, and share the UDN with others.
Conferences and Events
Sequencing Guide
RNA Sequencing Guide
MyGene2 Profiles
Contact Form